Não foi possível enviar o arquivo. Será algum problema com as permissões?
Exercícios
Semana 1
- Produce a plot of the Rongelap data in which a continuous colour scale or grey scale is used to indicate the value of the emission count per unit time at each location, and the two sub-areas with the 5 by 5 sub-grids at 50 metre spacing are shown as insets.
- Construct a polygonal approximation to the boundary of The Gambia. Construct plots of the malaria data which show the spatial variation in the values of the observed prevalence in each village and of the greenness covariate.
- (1) Consider the elevation data as a simple regression problem with elevation as the response and north-south location as the explanatory variable. Fit the standard linear regression model using ordinary least squares. Examine the residuals from the linear model, with a view to deciding whether any more sophisticated treatment of the spatial variation in elevation might be necessary.
- (2) Find a geostatistical data-set which interests you.
- What scientific questions are the data intended to address? Do these concern estimation, prediction, or testing?
- Identify the study region, the design, the response and the covariates, if any.
- What is the support of each response?
- What is the underlying signal?
- If you wished to predict the signal throughout the study region, would you choose to interpolate the response data?
- Load the Paraná data-set from geoR using the command
data(parana)
and inspect its documentation usinghelp(parana)
. For these data, consider the same questions as were raised in Exercise 1.4. - Read the Chapter 2 of Diggle & Ribeiro (2007) (you can get this chapter here)
Semana 2
- (3) load the data sets
parana,Ksateca20available ingeoRusing commands such as:data(parana)
and the documentation describing each data set with thehelp()functionhelp(parana)
Perform exploratory data analysis and build a model you find suitable for each data. - (3) In the examples above, would you have other candidate models for each data-set?
- Inspect an example geoestatistical analysis for the hydraulic conductivity data.
- (4) Consider the following two models for a set of responses,
associated with a sequence of positions
along a one-dimensional spatial axis
.
, where
and
are parameters and the
are mutually independent with mean zero and variance
.
where the
are as in (a) but A and B are now random variables, independent of each other and of the
, each with mean zero and respective variances
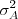 and
and  .
.
For each of these models, find the mean and variance ofand the covariance between
and
for any
. Given a single realisation of either model, would it be possible to distinguish between them?
- (5) Suppose that
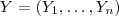 follows a multivariate Gaussian distribution with
follows a multivariate Gaussian distribution with 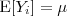 and
and 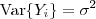 and that the covariance matrix of
and that the covariance matrix of can be expressed as
. Write down the log-likelihood function for
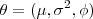 based on a single realisation of
based on a single realisation of and obtain explicit expressions for the maximum likelihood estimators of
and
when
is known. Discuss how you would use these expressions to find maximum likelihood estimators numerically when
is unknown.
- (6) Is the following a legitimate correlation function for a one-dimensional spatial process
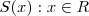 ? Give either a proof or a counter-example.
? Give either a proof or a counter-example.
- (7) Consider the following method of simulating a realisation of a one-dimensional spatial process on
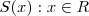 , with mean zero, variance 1 and correlation function
, with mean zero, variance 1 and correlation function . Choose a set of points
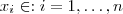 . Let
. Let denote the correlation matrix of
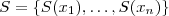 . Obtain the singular value decomposition of
. Obtain the singular value decomposition of as
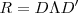 where
where is a diagonal matrix whose non-zero entries are the eigenvalues of
, in order from largest to smallest. Let
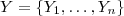 be an independent random sample from the standard Gaussian distribution,
be an independent random sample from the standard Gaussian distribution, 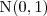 . Then the simulated realisation is
. Then the simulated realisation is 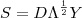
- (7) Write an
Rfunction to simulate realisations using the above method for any specified set of pointsand a range of correlation functions of your choice. Use your function to simulate a realisation of
on (a discrete approximation to) the unit interval
.
- (7) Now investigate how the appearance of your realisation
changes if in the equation above you replace the diagonal matrix
by truncated form in which you replace the last
eigenvalues by zeros.
Semana 3
- (8) Fit a model to the surface elevation data assuming a linear trend model on the coordinates and a Matérn correlation function with parameter
. Use the fitted model as the true model and perform a simulation study (i.e. simulate from this model) to compare parameter estimation based on maximum likelihood, restricted maximum likelihood and variograms.
- (9) Simulate 200 points in the unit square from the Gaussian model without measurement error, constant mean equals to zero, unit variance and exponential correlation function with
and anisotropy parameters
. Obtain parameter estimates (using maximum likelihood):
- assuming a isotropic model
- try to estimate the anisotropy parameters
Compare the results and repeat the exercise for.
- (10) Consider a stationary trans-Gaussian model with known transformation function
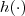 , let $x$ be an arbitrary
, let $x$ be an arbitrary
location within the study region and define . Find explicit expressions for
 where
where denotes the observed measurements on the untransformed scale and:
.
- (11) Analyse the Paraná data-set or any other data set of your choice assuming priors obtaining:
- a map of the predicted values over the area
- a map of the predicted std errors over the area
- a map of the probabilities of being above a certain (arbitrarily) chosen threshold over the area
- a map of the 10th, 25th, 50th, 75th and 90th percentiles over the area
- the predictive distribution of the proportion of the area with the value of the study variable below a certain threshold. (as a suggestion you can use the 30th percentile of the data as the value of such a threshold)
Semana 4
- (12) Consider the stationary Gaussian model in which
, where
is a stationary Gaussian process with mean zero, variance
and correlation function
, whilst the
are mutually independent
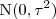 random variables. Assume that all parameters except
random variables. Assume that all parameters except are known. Derive the Bayesian predictive distribution of
for an arbitrary location
when
is assigned an improper uniform prior,
constant for all real
. Compare the result with the ordinary kriging formulae.
- (13) For the model assumed in the previous exercise, assuming a correlation function parametrised by a scalar parameter
obtain the posterior distribution for:
- a normal prior for
and assuming the remaining parameters are known
- a normal-scaled-inverse-
 prior for
prior for 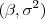 and assuming the correlation parameter is known
and assuming the correlation parameter is known - a normal-scaled-inverse-
prior for
and assuming a generic prior
for correlation parameter.
- (14) Analise the Paraná data-set or any other data set of your choice assuming priors for the model parameters and obtaining:
- the posterior distribution for the model parameters
- a map of the predictive mean over the area
- a map of the predictive median over the area
- the predictive distribution at three arbitrary selected locations within the area
- (15) Obtain simulations from the Poison model as shown in Figure 4.1 of the text book for the course.
- (15) Try to reproduce or mimic the results shown in Figure 4.2 of the text book for the course simulating a data set and obtaining a similar data-analysis. Note: for the example in the book we have used set.seed(34).
- (16) Reproduce the simulated binomial data shown in Figure 4.6. Use the package geoRglm in conjunction with priors of your choice to obtain predictive distributions for the signal
at locations
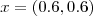 and
and 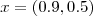 . Compare the predictive inferences which you obtained in the previous exercise with those obtained by fitting a linear Gaussian model to the empirical logit transformed data,
. Compare the predictive inferences which you obtained in the previous exercise with those obtained by fitting a linear Gaussian model to the empirical logit transformed data, . Compare the results of the two previous analysis and comment generally.
Semana 5
- (17) The composite likelihood (CL) is obtained by the product of independent distributions for pairs of variables at data locations. Assume a Gaussian model with constant mean and isotropic exponential correlation function.
- write down the expression of the CL and discuss how parameter estimates could be obtained
- write down a code to obtain CL parameter estimates for the s100 data set and compare with the ones given by ML and REML.